The collaborative study, now published in the Journal of Extracellular Vesicles, systematically measured the size, concentration, and particle size distribution of samples with varying complexity.
The work was driven by a critical need to develop reproducible and reliable characterisation protocols for the analysis of extracellular vesicles (EVs) – a need which is highly recognised by the EV and regulatory community. Here, we provide an overview of the study which compared:
- Nano flow cytometry (nFCM)
- Nanoparticle tracking analysis (NTA)
- Multi-angle dynamic light scattering (MADLS)
- Tunable resistive pulse sensing (TRPS)
- Centrifugal liquid sedimentation (CLS)
- Asymmetrical flow field flow fractionation coupled with multi-angle light scattering (AF4-MALS)
The study was conducted by researchers from The University of Queensland (Australia), Trinity College Dublin (Ireland), Izon Science (New Zealand), the National Research Council (CNR, Italy), NanoFCM (UK), Joint Research Centre (Italy), and SINTEF (Norway).
Analytical techniques put to the test using a four-stage, stepwise approach
To assess the capabilities and limitations of each analytical method, the group chose a selection of samples ranging in type and complexity. The simplest analytes were monomodal samples of NIST-traceable polystyrene particles, measuring 60, 100, 150, 200, and 240 nm in diameter. Then, to mimic the size distribution of an EV-containing plasma sample, bimodal, trimodal, and quadrimodal mixtures of polystyrene particles were assessed. Subsequent complexity was introduced via a commercially available, well-characterised PEGylated liposome sample. Finally, an EV-containing plasma sample was prepared using size exclusion chromatography. To avoid any subjective biases, instrument operators were blind to the sample composition of the monomodal and multimodal polystyrene samples.
Not all techniques made it past the second challenge
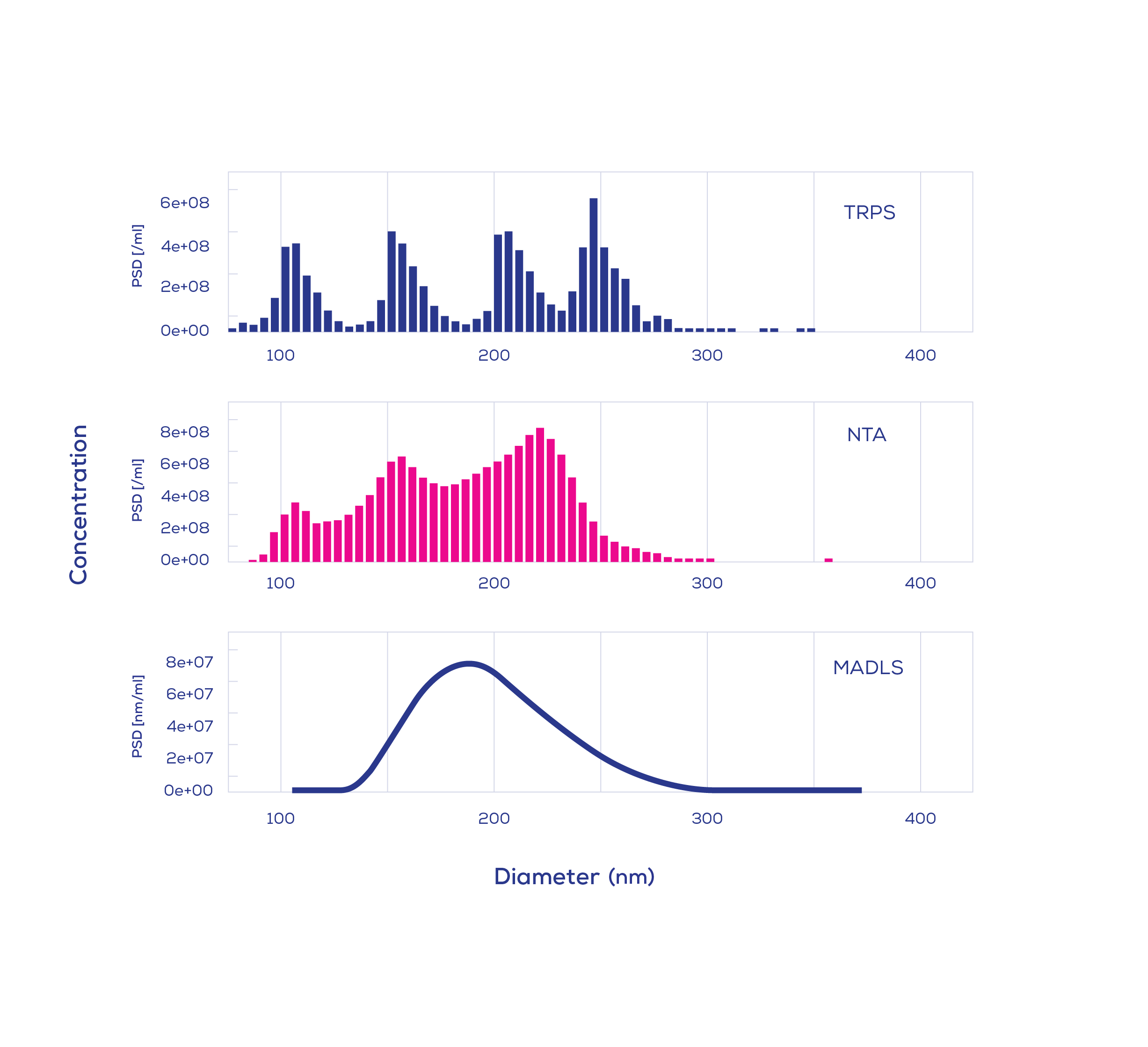
The use of known samples with incremental complexity enabled the performance of various platforms to be evaluated in a quantifiable way. Not all techniques could provide accurate size and concentration measurements, and their limitations were revealed at different levels of complexity. For monomodal samples, i.e., samples containing particles of the same size, all techniques performed similarly. In contrast, for bimodal mixtures (a 1:1 ratio of 100 and 200 nm particles), MADLS showed a broad distribution of particles and did not show any features representative of the multimodal composition.
Facing trimodal and quadrimodal challenges
When the analytical complexity was lifted to include trimodal and quadrimodal nanoparticle samples, further limitations were revealed. For instance, a trimodal mixture containing equal parts of 60, 100, and 150 nm particles could only be fully resolved using TRPS and nFCM. Instead of confirming a 1:1:1 ratio (with each particle size comprising 33.3% of the population), NTA underestimated the proportion of 60 nm particles to be 2-5% on multiple occasions. However, this phenomenon was not observed with larger particles (100, 150, and 200 nm); five of the six techniques (TRPS, nFCM, CLS, NTA, and AF4-MALS) were able to resolve this larger, trimodal mixture.
When faced with a quadrimodal mixture, only TRPS, CLS, and AF4-MALS clearly resolved the subpopulations. Given the extensive time commitment required to obtain CLS and AF4-MALS measurements, quadrimodal samples were characterised across one and two runs, respectively, while TRPS data was collected in triplicate. Meanwhile, NTA could only detect the presence of three subpopulations, none of which could be distinguished clearly. For nFCM, single measurements resolved quadrimodal mixtures, but a slight size-shift was observed across multiple runs. When nFCM data was collected and averaged from three runs, resolution was lost and the 200 and 240 nm peaks merged into one.
In total, the study included ten samples comprising polystyrene particles of varying sizes, ratios, and number of subpopulations. Each of the techniques measured with varying accuracy; for NTA, concentration measurements deviated from the nominal concentration by 60%. TRPS was at the other end of the spectrum, with concentration measurements falling within 10% of the nominal concentration.

Approaches validated with liposome analysis
To validate the protocols with traceable standards, Vogel et al. (2021) obtained a monodisperse, PEGylated liposome sample containing three different lipids. The number-based concentration of liposomes was calculated to be 2.14*1014 particles/mL using several theoretical assumptions. The calculated liposome number concentration was in good agreement with the mean concentration averaged across measurements taken with TRPS, NTA, and AF4-MALS – each of which calculated liposome size within 10% of its nominal diameter (78 nm).
In contrast, nFCM significantly underestimated liposome concentration (up to 11 times lower than other techniques) and produced a mode diameter of 53 nm. Of note, AF4-MALS was highly sensitive to the refractive index, which is difficult to calculate for non-uniform biological nanoparticles such as liposomes and EVs. For CLS, the difference in density between liposome particles and sucrose gradient was insufficient for analysis, highlighting the need to explore ultracentrifugation and higher centrifugal forces if CLS is to be used in this context.
Assessing performance using biological samples
In the final test, analytical techniques were assessed on their ability to measure the concentration and particle size distribution of complex biological samples, i.e., plasma samples containing EVs (80-250 nm and 50-400 nm). TRPS concentration measurements showed good repeatability, with the coefficient of variation below 10%. CLS concentration measurements were two orders of magnitude higher than values for TRPS, NTA and nFCM. EV concentration could not be obtained using AF4-MALS, despite the efforts of two laboratories who followed a recently published AF4-MALS protocol (Zhang & Lyden 2019)2. Given that MADLS failed to resolve a quadrimodal sample containing particles in the size range of EV isolates, MADLS measurements of EV isolates were not attempted.
Who is up to the challenge? A strong performance from TRPS
Sensitive, accurate, and reproducible methods for nanoparticle characterisation are needed to support work across many fields of biological and environmental research. For the EV community, which ultimately aims to bring EV-related applications to the clinic, size and concentration must be measured in a standardised way to assure quality, potency, stability and batch-to-batch consistency. To aid in the development of robust protocols, Vogel et al. (2021) present a comparative assessment of six analytical techniques applied to samples of varying type and complexity.
Of the six techniques, TRPS performed strongly, enabling reproducible size and concentration measurements to be obtained for polystyrene mixtures, liposomes, and EV isolates. Critically, TRPS provided a high level of resolution for polydisperse samples and produced size and concentration measurements that aligned with the nominal diameters of the various NIST-traceable standards. The experimental polystyrene mixtures were accurately reflected in TRPS size distribution analysis, even for mixtures containing the smallest particles (60 nm). Overall, Vogel et al. (2021)1 revealed the limitations of a range of techniques and like many others, they recommend the use of multiple orthogonal approaches – ideally using at least one non-optical or hyphenated technique. Given the results from this study, TRPS can continue to be viewed as a strong contender.
Get in touch now to learn how TRPS can support your research